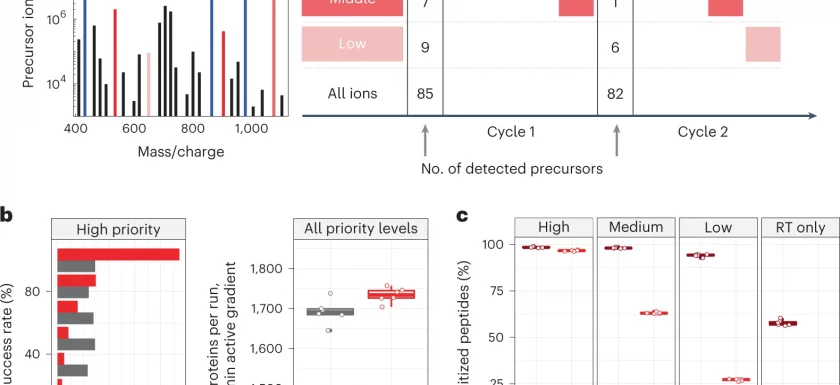
New paper: ‘Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics’ is now published in Nature Methods. In this paper led by Gray Huffman from the Slavov lab, MaxQuant.Live is applied for targeted analysis of single-cell proteomics experiments. A new prioritisation feature is used to permit within-duty-cycle selection of precursors of interest, improving run-to-run reproducibility, and mitigating data-dependent biases which favor analysis of more-intense ions, allowing improved sensitivity which can be applied to study post-translational modifications.
You can read the published paper here. The journal also released a research briefing on this work, which can be read here.
‘

Comments are closed, but trackbacks and pingbacks are open.