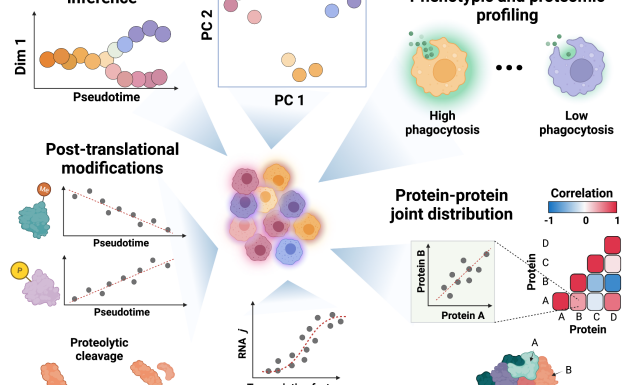
The lab contributed to a community effort to define standards for performing, benchmarking and reporting single-cell proteomics experiments. While SCP analysis has the potential to accurately quantify thousands of proteins across thousands of single cells, the accuracy and reproducibility of the results may be undermined by numerous factors affecting experimental design, sample preparation, data acquisition, and data analysis. Establishing community guidelines and standardized metrics will enhance rigor, data quality, and alignment between laboratories. Here we propose best practices, quality controls, and data reporting guidelines to assist in the broad adoption of reliable quantitative workflows for single-cell proteomics.
The preprint is available on Arxiv here: https://arxiv.org/abs/2207.10815

Comments are closed, but trackbacks and pingbacks are open.