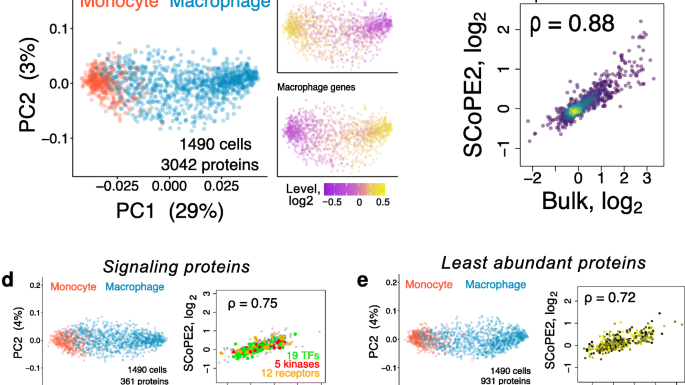
Our paper detailing the SCoPE2 method for single-cell proteomic analysis as applied to a cell culture model of macrophage heterogeneity is now published in genome biology. In this paper we revised the original SCoPE-MS method, generating a method capable of high-throughput analysis of single-cells with deep coverage. We manage to generate single-cell proteomes for 1490 single macrophage or monocyte cells, to a depth of 3042 proteins. You can read the paper here.

Comments are closed, but trackbacks and pingbacks are open.